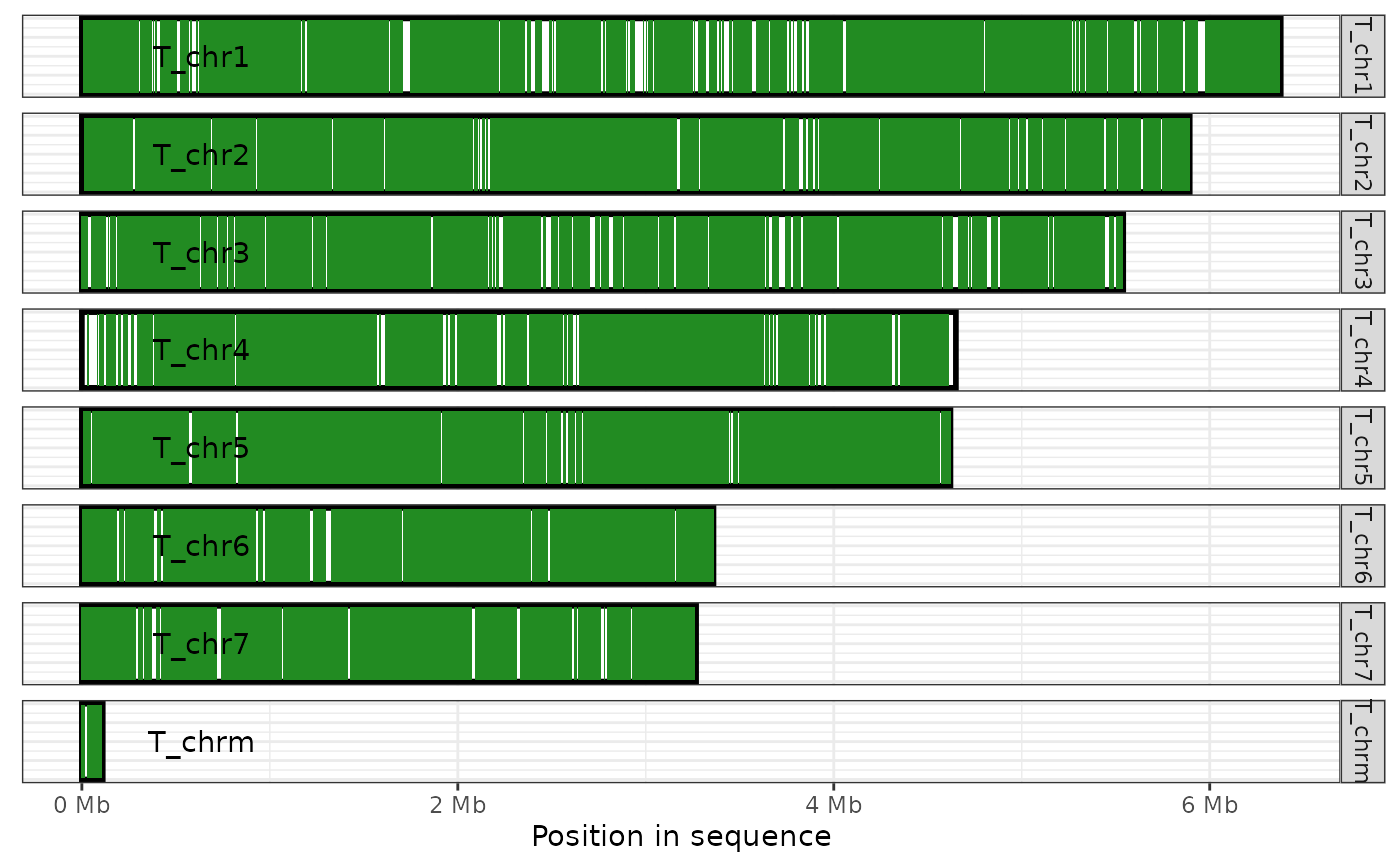
A minimalistic ggplot2 theme designed for use with genome coverage plots
Source:R/coverage_plot.r
theme_coverage_plot.RdThis theme is used as the default when plot_coverage is called,
so you should usually only call this function to modify the appearance of the
coverage plot.
theme_coverage_plot(facet_labs = TRUE, show_legend = TRUE)
Arguments
| facet_labs | logical If TRUE (default), label sequences using the facet
labels; if FALSE, sequences are labeled directly using
|
|---|---|
| show_legend | logical If TRUE (default), label display any legend
associated with the fill parameter of |
Examples
ali <- read_paf( system.file("extdata", "fungi.paf", package="pafr") ) plot_coverage(ali) + theme_coverage_plot(show_legend=FALSE)